SAM Angle Mapper Transform is a type of technique related to the Spectral Angle Mapper (SAM), a technique for comparing the similarity of two spectra by measuring the angle between them. It is often used for hyperspectral image analysis, such as target detection, classification, and endmember extraction. This article will show how to apply SAM to hyperspectral data using Python code examples and answer some frequently asked questions.
We will also introduce SAM Angle Mapper Transform, a specific function in the pci package that applies SAM to an image and reference spectrum files. SAM transform is similar to the SAM function but has some additional features and parameters. SAM transform can create new channels in the image file for the SAM angles and the classification results. It can also specify the input and output bands for SAM calculation, the SAM threshold for classification, and whether to output only the classified or all pixels.
This article will help you to understand and apply SAM and SAM Angle Mapper Transform to your hyperspectral data analysis tasks using Python. We will explain the concept, the syntax, and the examples of SAM and SAM Angle Mapper Transform and answer some frequently asked questions about them.
Contents
How to use SAM in Python?
There are several ways to use SAM in Python, depending on the package and function you choose. Here, we will show three examples using PySpTools, torchmetrics, and pci.
The formula for calculating the SAM angle between two spectra x and y is:
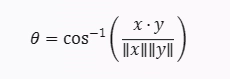
PySpTools
PySpTools is a Python package for hyperspectral image processing that provides various tools and algorithms, including SAM. To use SAM with PySpTools, install the package using pip or conda and import the SAM module. Then, you can use the SAM function to compute the SAM angle between an image cube and a reference spectrum. The function returns a numpy array of angles for each pixel in the image cube.
# Import modules
import numpy as np
import pysptools.sam as sam
# Load hyperspectral data
data_file = 'cuprite_subpix.hdr'
data_path = 'cuprite_subpix'
data_header = util.load_ENVI_header(data_file)
data_cube = util.load_ENVI_data(data_path, data_header['interleave'], data_header['data type'])
# Remove water vapor bands
wv_bands = [104, 105, 106, 107, 108, 120, 121, 122]
data_cube = np.delete(data_cube, wv_bands, axis=2)
# Select one endmember as reference spectrum
ref_spectrum = U[3,:]
# Compute SAM angle between image cube and reference spectrum
sam_angles = sam.sam(data_cube, ref_spectrum)torchmetrics
torchmetrics is a Python package for machine learning metrics that provides various functions for different domains, including image analysis. To use SAM with torchmetrics, you must install the package using pip or conda and import the SpectralAngleMapper class from torchmetrics.image module. Then you can create an instance of SpectralAngleMapper with a reduction method (such as elementwise_mean or sum), and call it with two tensors of predictions and targets. The function returns a tensor of SAM angles for each pair of spectra in the tensors.
# Import modules
import torch
import torchmetrics
# Generate random predictions and targets
torch.manual_seed(42)
preds = torch.rand([16, 3, 16, 16])
target = torch.rand([16, 3, 16, 16])
# Create an instance of SpectralAngleMapper with elementwise_mean reduction
sam = torchmetrics.image.SpectralAngleMapper(reduction='elementwise_mean')
# Compute SAM angle between predictions and targets
sam_angles = sam(preds, target)pci
pci is a Python package for geospatial image processing that provides various functions for different tasks, such as classification, segmentation, and transformation. To use SAM with pci, you must install the package using pip or conda, and import the SAM function from the pci.sam module. Then, you can use the SAM function to compute the SAM angle between an image file and a reference spectrum file. The function creates new channels for the SAM angles and classification results in the image file.
# Import modules
import pci.sam
# Specify input and output files
input_file = 'cupref.pix'
ref_file = 'ref04b.spl'
output_channels = list(range(226, 233 + 1)) # newly added channels for SAM angles
class_channel = 234 # newly added channel for classification
# Specify SAM threshold
sam_threshold = 0.1 # based on trial and error
# Compute SAM angle between image file and reference spectrum file
pci.sam.sam(fili=input_file,
dbib=[],
specfile=ref_file,
dboc=output_channels,
dbclass=class_channel,
samthres=sam_threshold,
valonly='yes')What is SAM Angle Mapper Transform?
SAM Angle Mapper Transform is a function in the pci package that applies SAM to an image file and a reference spectrum file. It is similar to the sam function but has some additional features and parameters. SAM transform can create new channels in the image file for the SAM angles and the classification results. It can also specify the input and output bands for SAM calculation, the SAM threshold for classification, and whether to output only the classified or all pixels.
The syntax for SAM Angle Mapper Transform is:
pci.sam.sam(fili, dbib, specfile, dboc, dbclass, samthres, valonly)where:
- fili is the input image file name.
- dbib is a list of input bands to use for SAM calculation. If empty, all bands are used.
- specfile is the reference spectrum file name.
- dboc is a list of output channels to store the SAM angles. The number of channels must match the number of spectra in the reference file.
- dbclass is the output channel to store the classification results. If zero, no classification is performed.
- samthres is the SAM threshold for classification. If zero, the default value of 0.1 is used.
- valonly is a flag to indicate whether to output only the classified pixels or all pixels. If ‘yes’, only the classified pixels are output. If ‘no’, all pixels are output.
To use SAM Angle Mapper Transform in Python, you must install the pci package using pip or conda and import the SAM function from the pci.sam module. Then, you can use the SAM function with the appropriate parameters to apply SAM to your image and reference spectrum files. The function will create new channels in your image file for the SAM angles and the classification results. You can then use other functions from pci to display or manipulate the results.
Here is an example code using SAM Angle Mapper Transform in Python:
# Import modules
import pci.sam
# Specify input and output files
input_file = 'cupref.pix'
ref_file = 'ref04b.spl'
output_channels = list(range(226, 233 + 1)) # newly added channels for SAM angles
class_channel = 234 # newly added channel for classification
# Specify SAM threshold
sam_threshold = 0.1 # based on trial and error
# Apply SAM transform to image file and reference spectrum file
pci.sam.sam(fili=input_file,
dbib=[],
specfile=ref_file,
dboc=output_channels,
dbclass=class_channel,
samthres=sam_threshold,
valonly='yes')FAQs
What are the advantages and disadvantages of SAM?
The advantages of SAM are that it is simple, fast, robust to noise and illumination variations, and does not require any prior knowledge or assumptions about the data. The disadvantages of SAM are that it is sensitive to spectral resolution and band selection and may not capture the spectral variability within a class or between classes.
How do we choose a suitable SAM threshold for classification?
This question has no definitive answer, as it depends on the data and the application. A common way to choose a SAM threshold is to try different values and compare the results visually or quantitatively. Alternatively, some methods have been proposed to determine the optimal SAM threshold automatically based on statistical criteria or optimization techniques.
How do we compare SAM with other spectral similarity measures?
There are many other spectral similarity measures besides SAM, such as Spectral Correlation Mapper (SCM), Spectral Information Divergence (SID), Euclidean Distance (ED), Mahalanobis Distance (MD), etc. Each measure has advantages and disadvantages and may perform differently depending on the data and the application. A comprehensive comparison of different spectral similarity measures can be found in this paper.
Conclusion
This article shows how to use SAM in Python using three different packages and functions: PySpTools, torchmetrics, and pci. We have also introduced SAM transform, a function in the pci package that applies SAM to an image file and a reference spectrum file. We have also answered some frequently asked questions about SAM. We hope this article has been helpful for you to apply SAM to your hyperspectral data analysis tasks.
Reference
To learn more about python related errors and modules follow Python Clear.

